Researchers Test Large Language Model that Preserves Patient Privacy
Performance of free, publicly available LLM is similar to ChatGPT for some tasks

Locally run large language models (LLMs) may be a feasible option for extracting data from text-based radiology reports while preserving patient privacy, according to a new study from the National Institutes of Health Clinical Center (NIH CC) published in Radiology.
Recently released LLM models such as ChatGPT and GPT-4 have garnered attention. However, they are not compatible with health care data due to privacy constraints.
“ChatGPT and GPT-4 are proprietary models that require the user to send data to OpenAI sources for processing, which would require de-identifying patient data,” said senior author Ronald M. Summers, MD, PhD, senior investigator in the Radiology and Imaging Sciences Department at the NIH. “Removing all patient health information is labor-intensive and infeasible for large sets of reports.”
In this study, led by Pritam Mukherjee, PhD, staff scientist at the NIH CC, researchers tested the feasibility of using a locally run LLM, Vicuna-13B, to label key findings from chest X-ray reports from the NIH and the Medical Information Mart for Intensive Care (MIMIC) Database, a publicly available dataset of de-identified electronic health records.
“Preliminary evaluation has shown that Vicuna, a free publicly available LLM, approaches the performance of ChatGPT in tasks such as multi-lingual question answering,” Dr. Summers said.
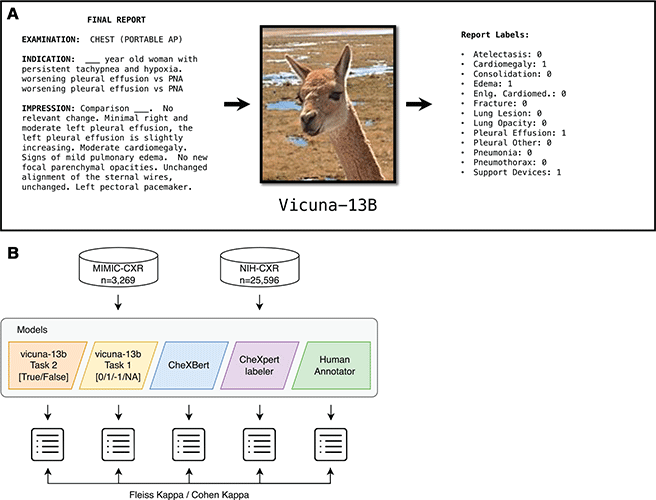
Overview of the study. (A) The open-access large language model Vicuna-13B, which can be run on a local computer without the need for de-identification of patient data, was prompted to examine unstructured, free-text chest radiography (CXR) reports and generate an output file reporting the results of 13 specific findings. AP = anteroposterior, Enlg. Cardiomed. = enlarged cardiomediastinum, PNA = pneumonia. (B) Reports from the MIMIC-CXR data set (n = 3269) and the National Institutes of Health (NIH) data set (n = 25 596) were used in this study. Vicuna was given two independent tasks that generated two different output files, one in which the 13 possible findings were labeled as positive or negative (task 2, orange model) and the other in which the 13 possible findings were labeled as positive, negative, unsure, or not mentioned (task 1, yellow model). The agreement between Vicuna model outputs and the CheXbert labeler, CheXpert labeler, and human annotations were compared using Fleiss or Cohen κ as appropriate.
https://doi.org/10.1148/radiol.231147 ©RSNA 2023LLM Tools Useful for Feature Extraction
The study dataset included 3,269 chest X-ray reports obtained from MIMIC and 25,596 reports from the NIH.
Using two prompts for two tasks, the researchers asked the LLM to identify and label the presence or absence of 13 specific findings on the chest X-ray reports. Researchers compared the LLM’s performance with two widely used non-LLM labeling tools.
A statistical analysis of the LLM output showed moderate to substantial agreement with the non-LLM computer programs.
“Our study demonstrated that the LLM’s performance was comparable to the current reference standard,” Dr. Summers said. “With the right prompt and the right task, we were able to achieve agreement with currently used labeling tools.”
Dr. Summers said LLMs that can be run locally will be useful in creating large data sets for AI research without compromising patient privacy.
“LLMs have turned the whole paradigm of natural language processing on its head,” he said. “They have the potential to do things that we've had difficulty doing with traditional pre-large language models.”
Listen as Dr. Summers discusses his research on testing large language models that preserve patient privacy.
Dr. Summers said LLM tools could be used to extract important information from other text-based radiology reports and medical records, and as a tool for identifying disease biomarkers.
“My lab has been focusing on extracting features from diagnostic images,” he said. “With tools like Vicuna, we can extract features from the text and combine them with features from images for input into sophisticated AI models that may be able to answer clinical questions.
“LLMs that are free, privacy-preserving, and available for local use are game changers,” he said. “They're really allowing us to do things that we weren't able to do before.”
For More Information
Access the Radiology study, “Feasibility of Using the Privacy-preserving Large Language Model Vicuna for Labeling Radiology Reports,” and the related editorial, “Feasibility and Prospect of Privacy-preserving Large Language Models in Radiology.”
Read previous RSNA News stories about large language models:
- ChatGPT Passes Radiology Board Exam
- Preparing Radiology Trainees for AI and ChatGPT
- The Good, the Bad and the Ugly of Using ChatGPT